
UMBC researchers used a transmission electron microscope to capture images of a satellite virus attaching to a helper virus. (Image Credit: Tagide deCarvalho)
Some viruses (satellites) typically rely on the host organism and another virus (helper) to complete its life cycle. This helper is essential for the virus to replicate its DNA or build the protective shell called capsid that shelters the genetic material. Scientists usually see these viruses close to each other but haven’t recorded a satellite latching on a helper before. For the first time, UMBC scientists used a transmission electron microscope (TEM) to discover a bacteriophage attaching itself to a helper virus.
The team’s discovery started as a project in the SEA-PHAGES program, which involved students using the Promega Wizard DNA purification system to isolate bacteriophages from environmental samples. They then sent them to the Pittsburgh Bacteriophage Institute for sequencing and analyzed the results using bioinformatics tools. One sample contained a MindFlayer DNA virus, and analysis revealed that unknown DNA contaminated it. Sequencing was achieved with the MiSeq sequencing platform using the NEB Ultra II Library Kit and 150-base single-end reads.
The team used the Morgagni M268 40 – 100kV TEM with an Orius CCD Camera mounted on a 35mm port or post column-wise (exchangeable) to look at the samples more closely. Specific software controls the imaging and analysis. Running on a Dual Pentium PC using Microsoft Windows 2000, this TEM comes with a user interface that has six fixed set-up pages (vacuum, high tension, column, stage, camera, and alignment). Digital images are captured via the user interface, which also means that third-party software can be taken over by the microscope for quicker and easier image acquisition.
The microscope’s magnification (full range of 25-200000 x) system focuses energy loss electrons and no loss electrons to deliver sharp, high-contrast images. Optimal image quality can be achieved thanks to the user-selectable automatic contrast enhancement (ACE) and a wobbler focus adjustable in direction and amplitude. The microscope features a vacuum system for pumping via a Diffstak TM pumping assembly. It comes with a buffer tank, so users won’t need to operate the pre-vacuum pump constantly. This has a working pressure of 5 x 10-3 Pa (5 x 10-5 Torr) and an ultimate pressure of < 1 x 10-3 Pa (< 10-5 Torr).
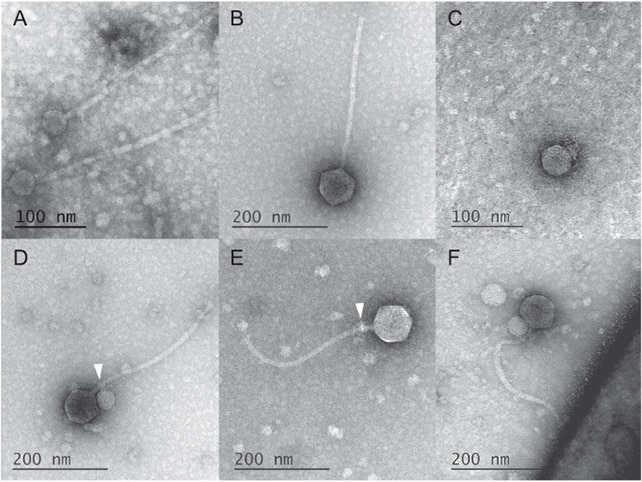
TEM images of the satellite and helper virus. The image in D shows both latching onto each other. (Image Credit: deCarvalho et al., The ISME Journal, 2023)
The team placed crude lysate (10 µL) atop 200 mesh formvar-covered, carbon-coated carbon grids. Afterward, they incubated it for one minute before rinsing it with ultra-pure water and staining it with 2% uranyl acetate for two minutes. According to the paper, scanning the grids enables the recording of the relevant satellite viruses’ number and position for the helper viruses. A motor stage stores the XY coordinates of the virus’ positions, and quick examination is often achieved through the special exposure select function.
X-Y coordinates, illumination, magnifications, and focus are selected before the automatic exposure sequence. Once the exposure button is pressed, the microscope moves, changes magnification, illumination, and focus, and moves the motor stage to the proper position. Users can repeat these steps until all the desired positions are imaged.
“For certain analyses noted in the results, we also used “picked” plaque samples that are prepared by placing 50 µL of phage buffer directly onto a plaque and incubating for 30 seconds before pipetting up the liquid, followed by staining methods described above for the crude lysate,” the team wrote in the paper.
They noticed the MiniFlayer (satellite virus) attached to the neck of the MindFlayer helper. Their images reveal that a satellite latched to 80% helpers at the neck. But those without the attachments had satellite tendrils, suggesting they were previously attached.
They also analyzed the host, satellite, and helper viruses’ genomes and noticed that MiniFlayer didn’t have the gene that helps it integrate into the host’s DNA. This suggests the virus has to stay near the MindFlayer helper to replicate within the host cell.
Have a story tip? Message me at: http://twitter.com/Cabe_Atwell

-

DAB
-
Cancel
-
Vote Up
0
Vote Down
-
-
Sign in to reply
-
More
-
Cancel
Comment-

DAB
-
Cancel
-
Vote Up
0
Vote Down
-
-
Sign in to reply
-
More
-
Cancel
Children