I saw this post by Shane Fleming on the PYNQ discussion board that I thought would be of interest to the PYNQ users in the E14 community: Interactive C++ on the Kria SoM in Jupyter Lab
It caught my eye because up to this point I haven't tried interactive C++ with Jupyter notebooks although I have heard about the Cling implementation. The post also demonstrates how to run pre-existing Vitis AI C++ code from the Vitis AI library in a notebook on the Kria SoM.
Shane's post covers the implementation steps in detail, so I'll just highlight them.
- Start with the PYNQ Ubuntu image on the Kria Starter Kit
- Install Miniconda and Mamba package managers to enable the installation of xeus-cling and PYNQ packages
- Installation should take about 30 minutes - unfortunately I ran into an unattended Ubuntu upgrade - so it took me more than an hour (need to remember to disable that)
- After that just need to start the Jupyter server and open the browser on port 9090 of the Kria
If everything has installed correctly you should see C++ notebook options in the launcher:
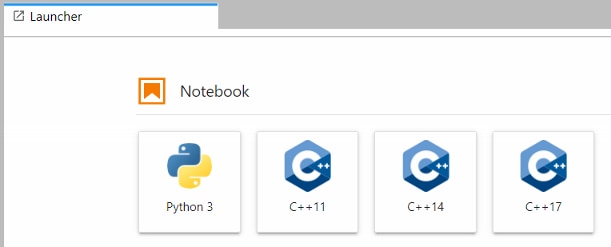
I'm primarily interested in the DPU-PYNQ example, so I went directly to that. A cpp_demo directory has been added under the pynq-dpu notebooks directory
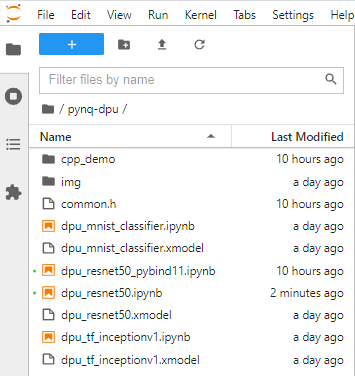
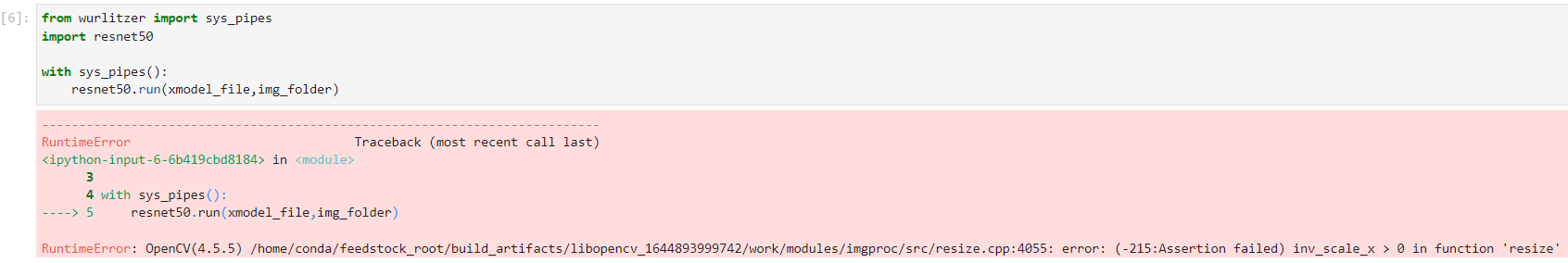
The example is going to run the resnet50 model using the pre-existing C++ code rather than using the Python bindings (pybind11) to run the C++ code in the previous example dpu_resnet50_pybind11 notebook.
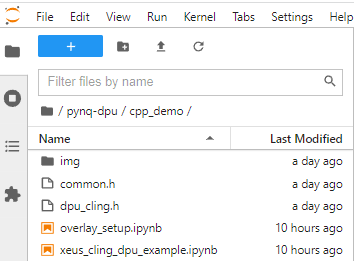
Two notebooks are being used:
- A Python notebook to load overlays
- A C++ notebook to run the application code
I've actually never run notebooks side-by-side, so another new experience. It's quite a bit faster than the pybinding code which takes about 30 seconds to compile - so it feels almost instantaneous.
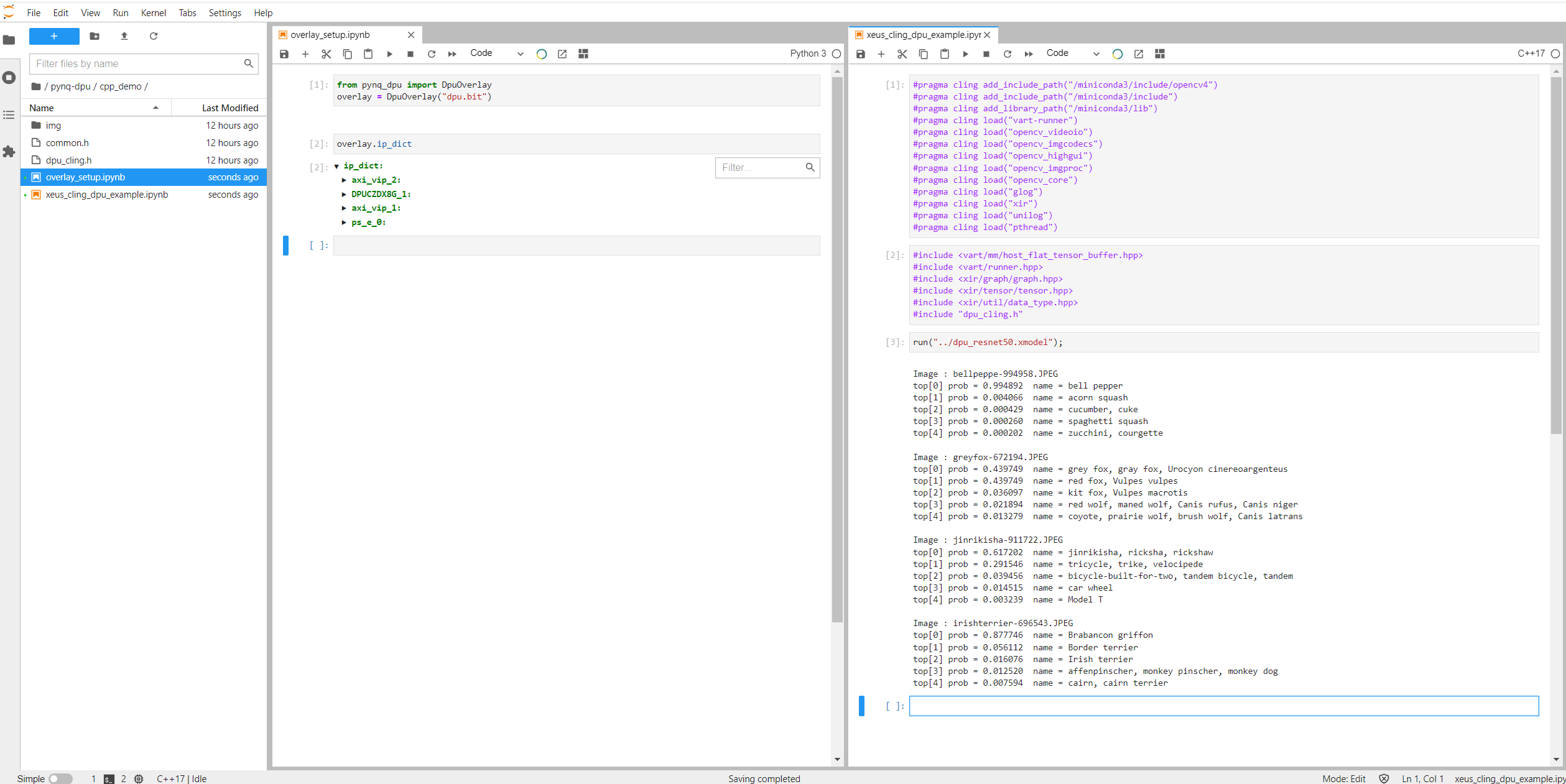
I did notice that the new PYNQ installation did break the original pybind11 notebook. I reported that to Shane and he was going to see if it is an OpenCV version issue.
It will be interesting to try some of the other models.
